Artificial Intelligence (AI) Assisted Detection of FGFR3 Alterations In Bladder Cancer From Scanned Whole Slide Images (WSI) of H&E Sections
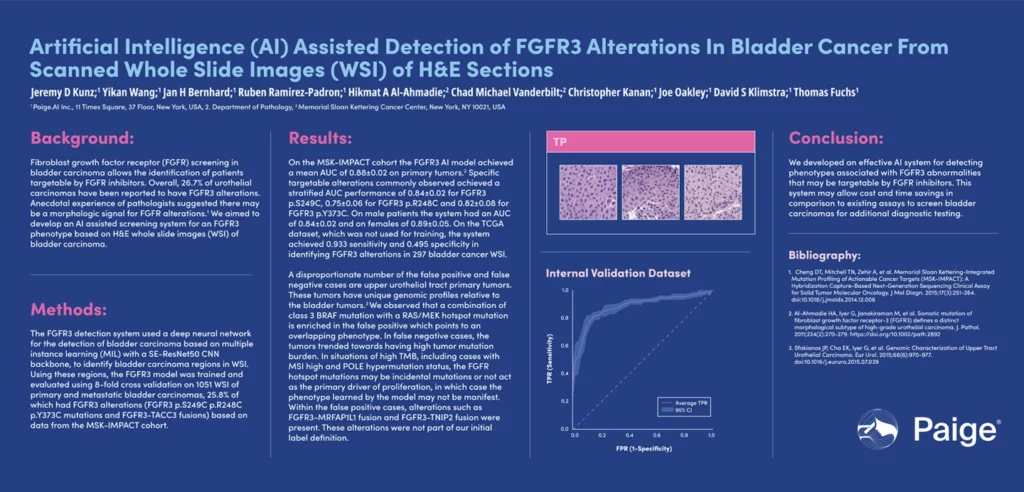
Paige developed an effective AI system for detecting phenotypes associated with FGFR3 abnormalities that may be targetable by FGFR inhibitors. This system may allow cost and time savings in comparison to existing assays to screen bladder carcinomas for additional diagnostic testing.
Artificial Intelligence (AI) Image Analysis for Chromosomal Instability (CIN) in Primary and Metastatic Breast Cancers (BC)
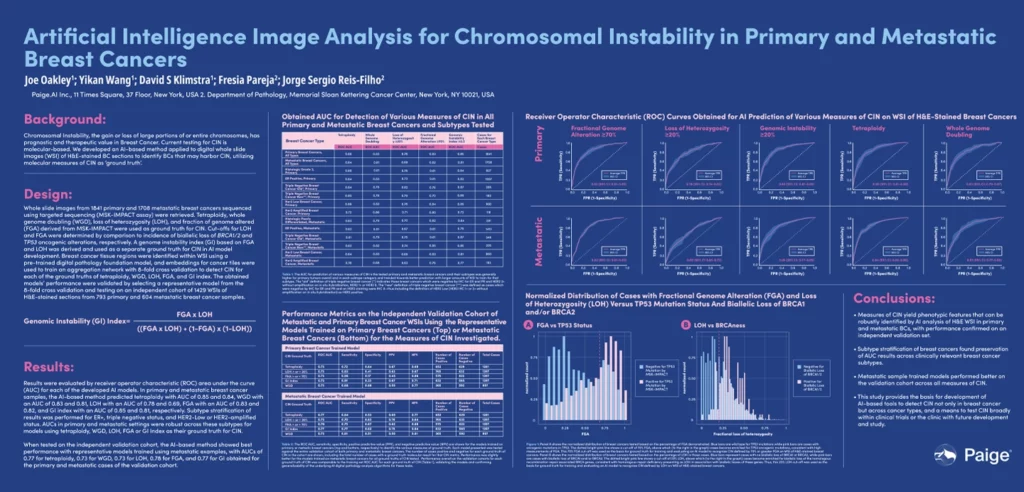
Measures of CIN yield phenotypic features that can be robustly identified by AI analysis of H&E WSI in primary and metastatic BCs. Subtype stratification of BC was robust with respect to AUC and more readily detectable in primary than in metastatic BC. This study provides the basis for development of AI-based tools to detect CIN not only in breast cancer but across cancer types, and a means to test CIN broadly within clinical trials.
Artificial Intelligence (AI) Assisted Detection of Microsatellite Instability Mismatch Repair Deficiency (MSI-H dMMR) in Multiple Tumor Types from Whole Slide Images (WSI) of H&E Sections
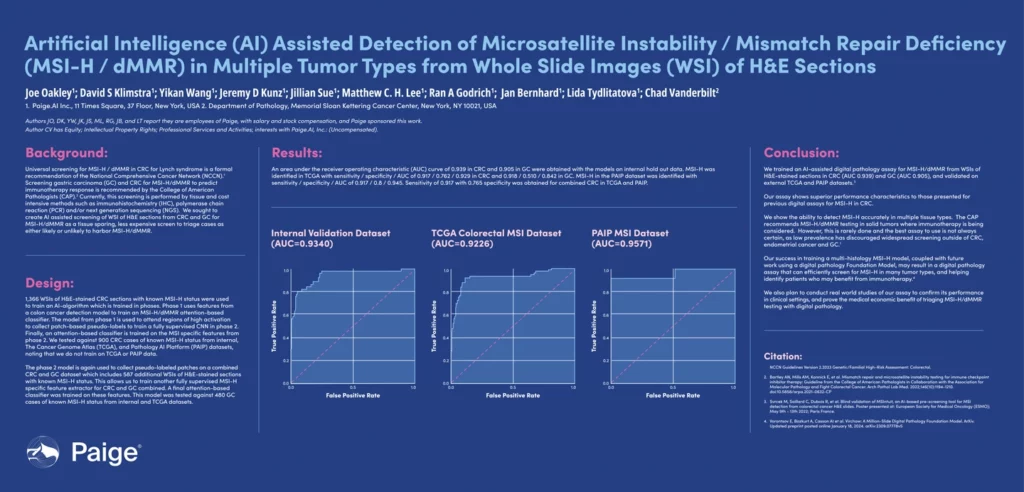
Paige trained an AI-assisted digital pathology assay for MSI-H/dMMR from WSIs of H&E-stained sections in CRC (AUC 0.939) and GC (AUC 0.905). Validation in CRC and GC from TCGA and PAIP datasets confirmed generalization to unseen external data. These results compare favorably to prior digital assays for MSI-H in CRC and GC.
The Reading Paradigm: How the Sequence and Presentation of AI Results to Pathologists Influences Endpoints and Outcomes

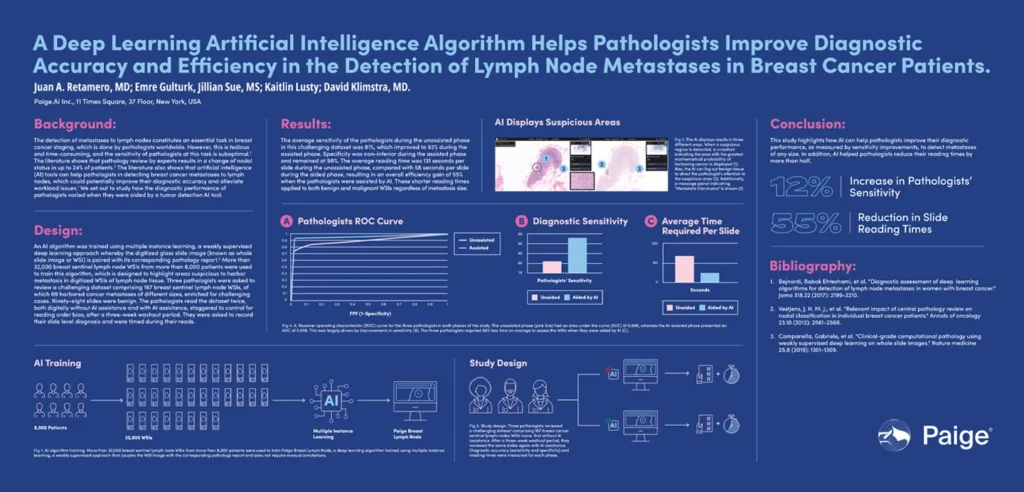
A Deep Learning Artificial Intelligence Algorithm Helps Pathologists Improve Diagnostic Accuracy And Efficiency In The Detection Of Lymph Node Metastases in Breast Cancer Patients
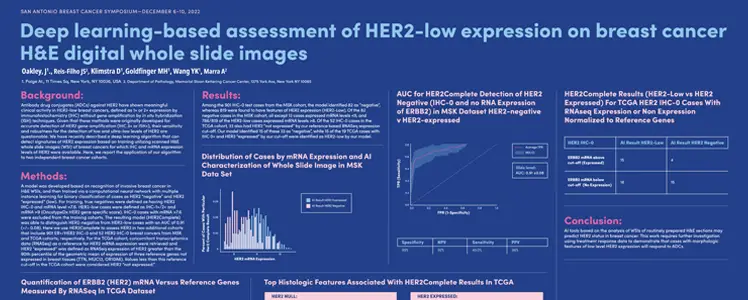
Deep learning-based assessment of HER2-low expression on breast cancer H&E digital whole slide images
High-throughput computational assessment of clinically relevant prostate cancer genetic phenotypes using AI analysis of H&E whole slide images
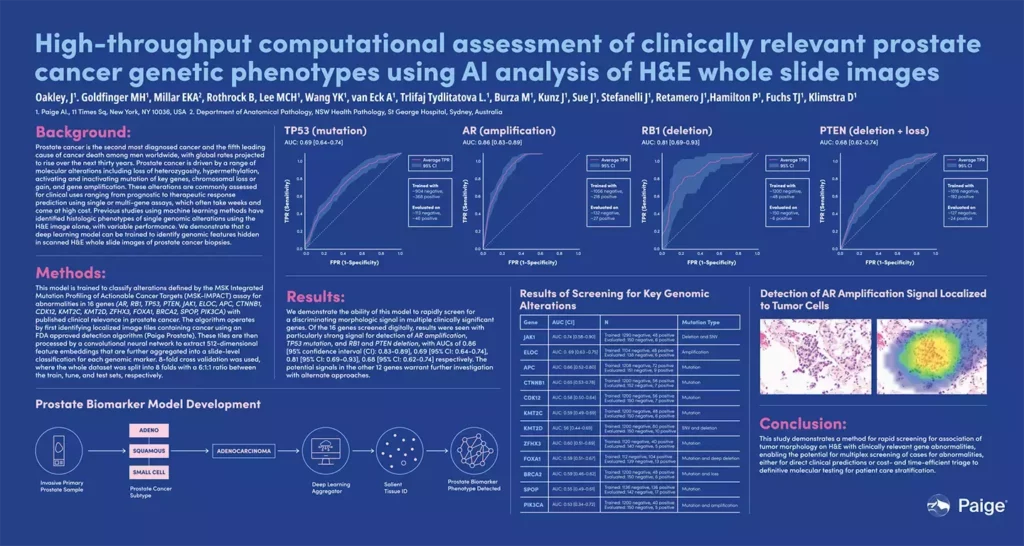
Oakley, J., Goldfinger, MH., Millar, EKA., Rothrock, B., Lee, MCH., Wang YK., van Eck, A., Trlifaj Tydilitatova, L., Burza, M., Kunz, J., Sue, J., Stefanelli, J., Retamero, J., Hamilton, P., Fuchs, TJ., Klimstra, D.
An AI-driven computational biomarker model from H&E slides recovers cases with low levels of HER2 from immunohistochemically HER2-negative breast cancers
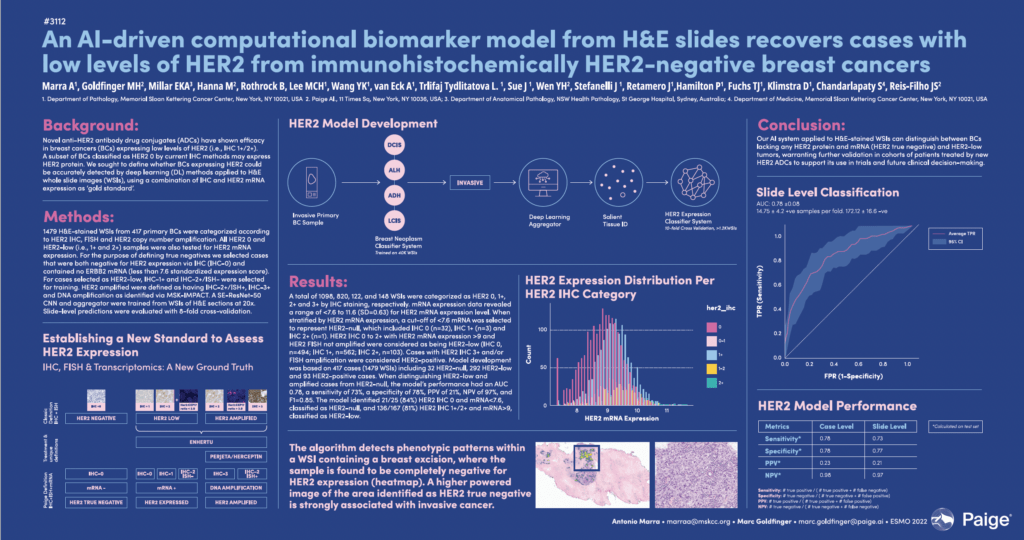
Marra A, Goldfinger M, Millar E, Hanna M, Rothrock B, Lee M, Wang Y, van Eck A, Trlifaj Tydlitatova L, Sue J, Wen Y, Stefanelli J, Retamero J,Hamilton P, Fuchs T, Klimstra D, Chandarlapaty S, Reis-Filho J
An Artificial Intelligence-Based Predictor of CDH1 Biallelic Mutations and Invasive Lobular Carcinoma
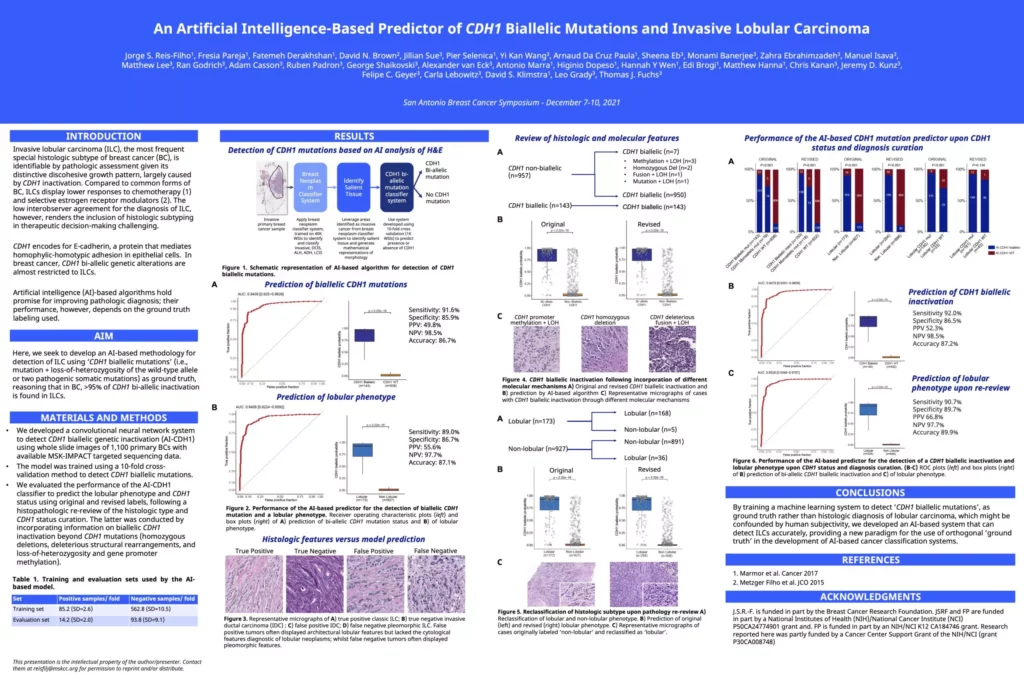
Jorge S. Reis-Filho, Fresia Pareja, Fatemeh Derakhshan, David N. Brown
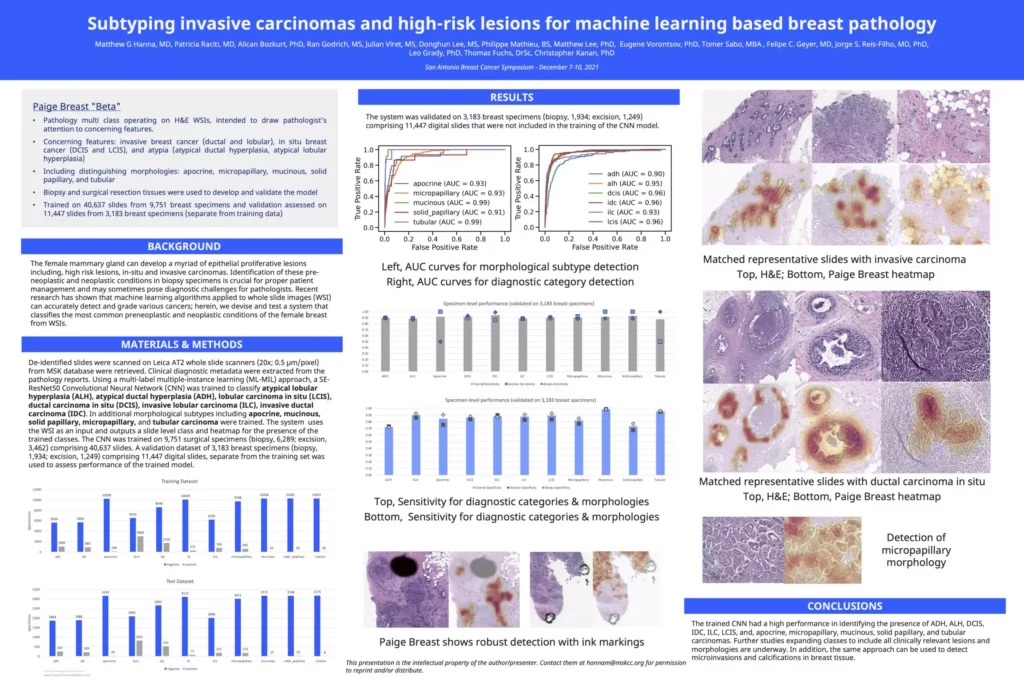
Subtyping invasive carcinomas and high-risk lesions for machine learning based breast pathology
Matthew G Hanna, MD, Patricia Raciti, MD, Alican Bozkurt, PhD, Ran Godrich, MS
